
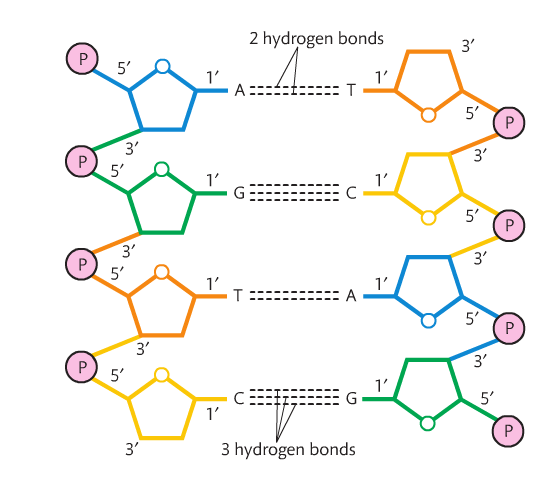
We report on the application of quantum chemical computational methods, in conjunction with spatial constraints derived from the experimental structure of a homopyrimidine PNA♽NA-PNA hetero-triplex, to investigate the influence of linker flexibility on binding interactions of the E-base with thymine and uracil bases in geometry-optimised model systems. An E-base (3-oxo-2,3-dihydropyridazine), attached to the polyamide backbone of a PNA Hoogsteen strand by a side-chain linker molecule, is typically used in the hydrogen bond recognition of the 4-oxo group of thymine and uracil nucleic acid bases in the major groove. Sequence-specific targeting of double-stranded DNA and non-coding RNA via triple-helix-forming peptide nucleic acids (PNAs) has attracted considerable attention in therapeutic, diagnostic and nanotechnological fields. These results are consistent with atomic force microscope measurements and provide the first theoretical validation of nearest neighbor approaches for predicting thermodynamic data of arbitrary DNA sequences. Our results indicate that (i) the effect of non-neighboring bases on the inter-strand interaction is negligibly small, (ii) London dispersion effects are essential for the stability of the duplex structure, (iii) the largest contribution to the stability of the duplex structure is the Watson-Crick base pairing - consistent with previous computational investigations, (iv) the effect of stacking between adjacent bases is relatively small but still essential for the duplex structure stability and (v) there are no cooperativity effects between intra-strand stacking and inter-strand base pairing interactions. Herein, we provide new insights into the intermolecular interactions responsible for the intrinsic stability of the duplex structure of a large portion of human B-DNA by using advanced quantum mechanical methods. Although not apparent from the current literature showing limited overlaps between the QM, simulation and bioinformatics studies of the nucleic acids backbone, there in fact should be a major cooperative interaction between these three approaches in studies of the sugar-phosphate backbone. The present status of the research is then illustrated by selected examples which include classification of DNA and RNA backbone families, benchmark structure-energy quantum chemical calculations, parameterization of the dihedral space of simulation force fields, incorporation of arsenate into DNA, sugar-phosphate backbone self-cleavage in small RNA enzymes, and intricate geometries of the backbone in recurrent RNA building blocks. We highlight main features, advantages and limitations of these techniques, with a special emphasis given to their synergy. We provide a side by side overview of structural biology/bioinformatics, quantum chemical and molecular mechanical/simulation studies of the nucleic acids backbone. Knowledge of geometrical and physico-chemical properties of the sugar-phosphate backbone substantially contributes to the comprehension of the structural dynamics, function and evolution of nucleic acids.


 0 kommentar(er)
0 kommentar(er)
